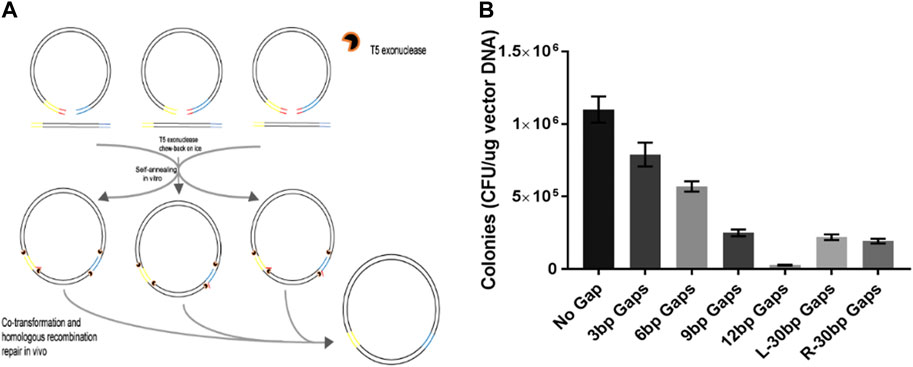
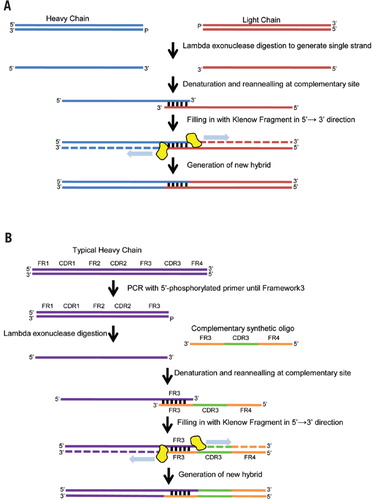
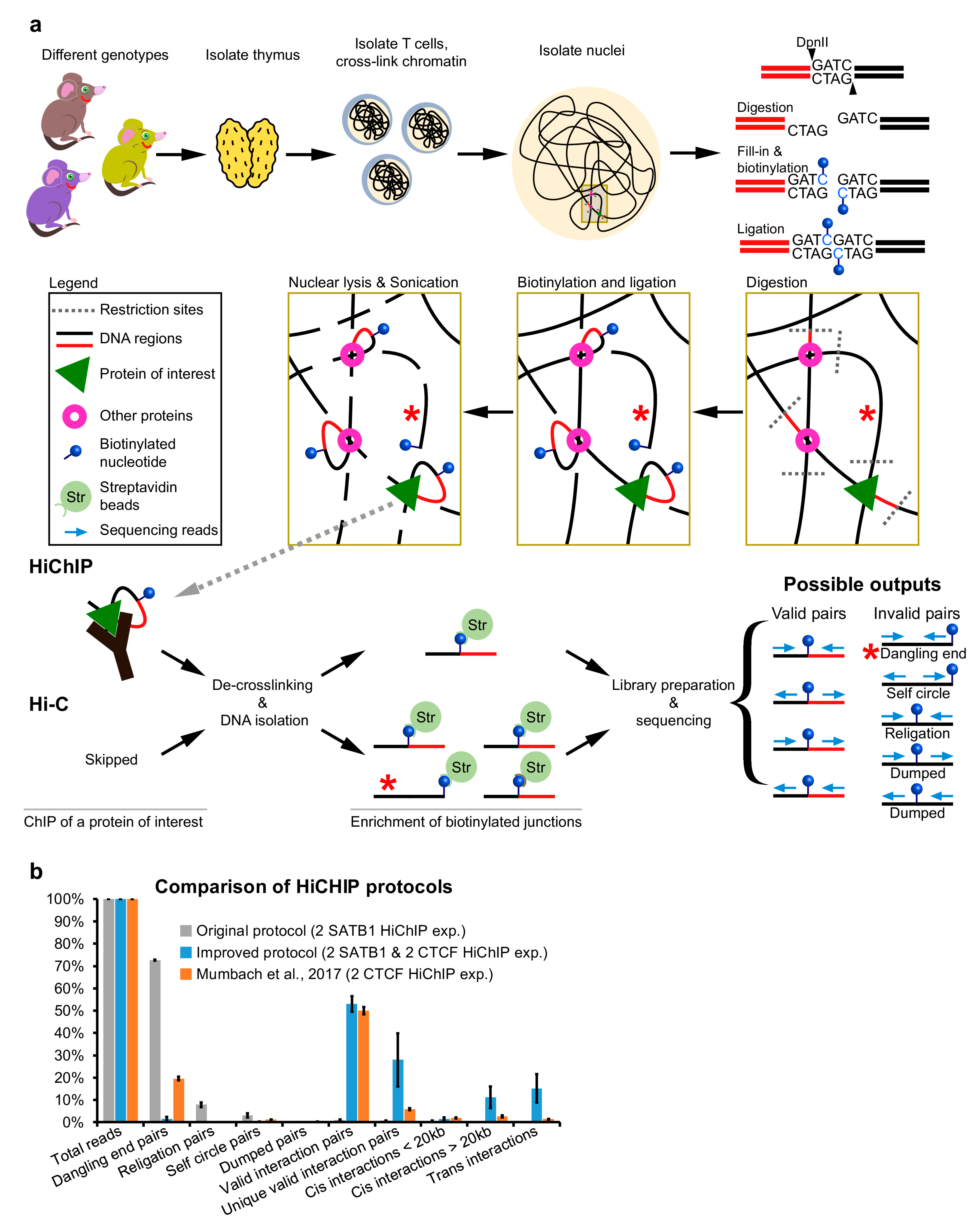
Weak Strand Displacement Activity Enables Human DNA Polymerase β to Expand CAG/CTG Triplet Repeats at Strand Breaks - ScienceDirect

Escherichia coli β-clamp slows down DNA polymerase I dependent nick translation while accelerating ligation | bioRxiv

WO1993000447A1 - Amplification of target nucleic acids using gap filling ligase chain reaction - Google Patents

Structural Transformation of Wireframe DNA Origami via DNA Polymerase Assisted Gap-Filling | ACS Nano

Structural Transformation of Wireframe DNA Origami via DNA Polymerase Assisted Gap-Filling | ACS Nano

Escherichia coli β-clamp slows down DNA polymerase I dependent nick translation while accelerating ligation | bioRxiv

A multifunctional DNA polymerase I involves in the maturation of Okazaki fragments during the lagging‐strand DNA synthesis in Helicobacter pylori - Cheng - 2021 - The FEBS Journal - Wiley Online Library

β-clamp inhibits strand displacement activity of exo-Klenow and exo-Pol... | Download Scientific Diagram

Structure of the Herpes Simplex Virus 1 Genome: Manipulation of Nicks and Gaps Can Abrogate Infectivity and Alter the Cellular DNA Damage Response | Journal of Virology
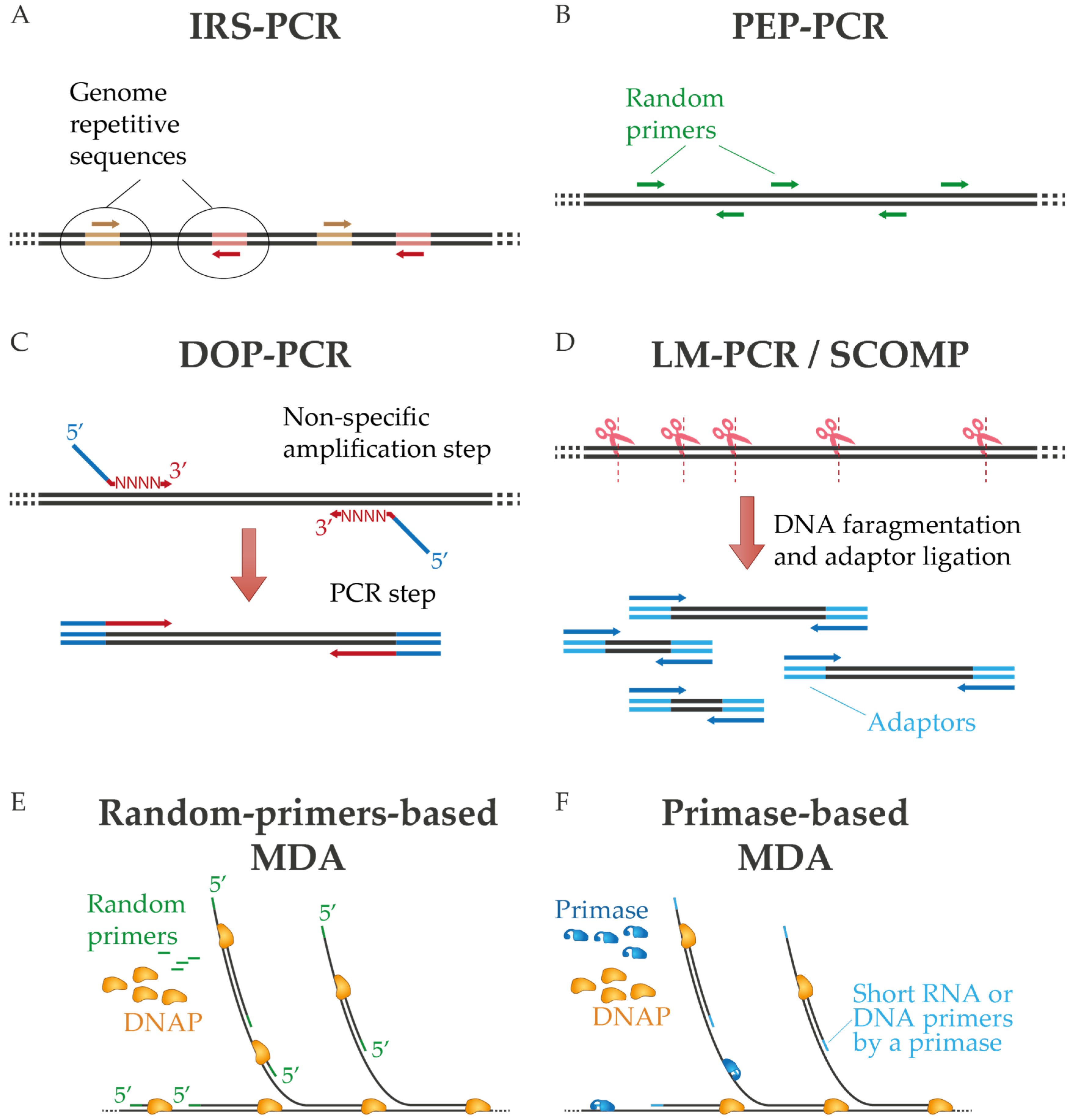
IJMS | Free Full-Text | DNA Polymerases for Whole Genome Amplification: Considerations and Future Directions
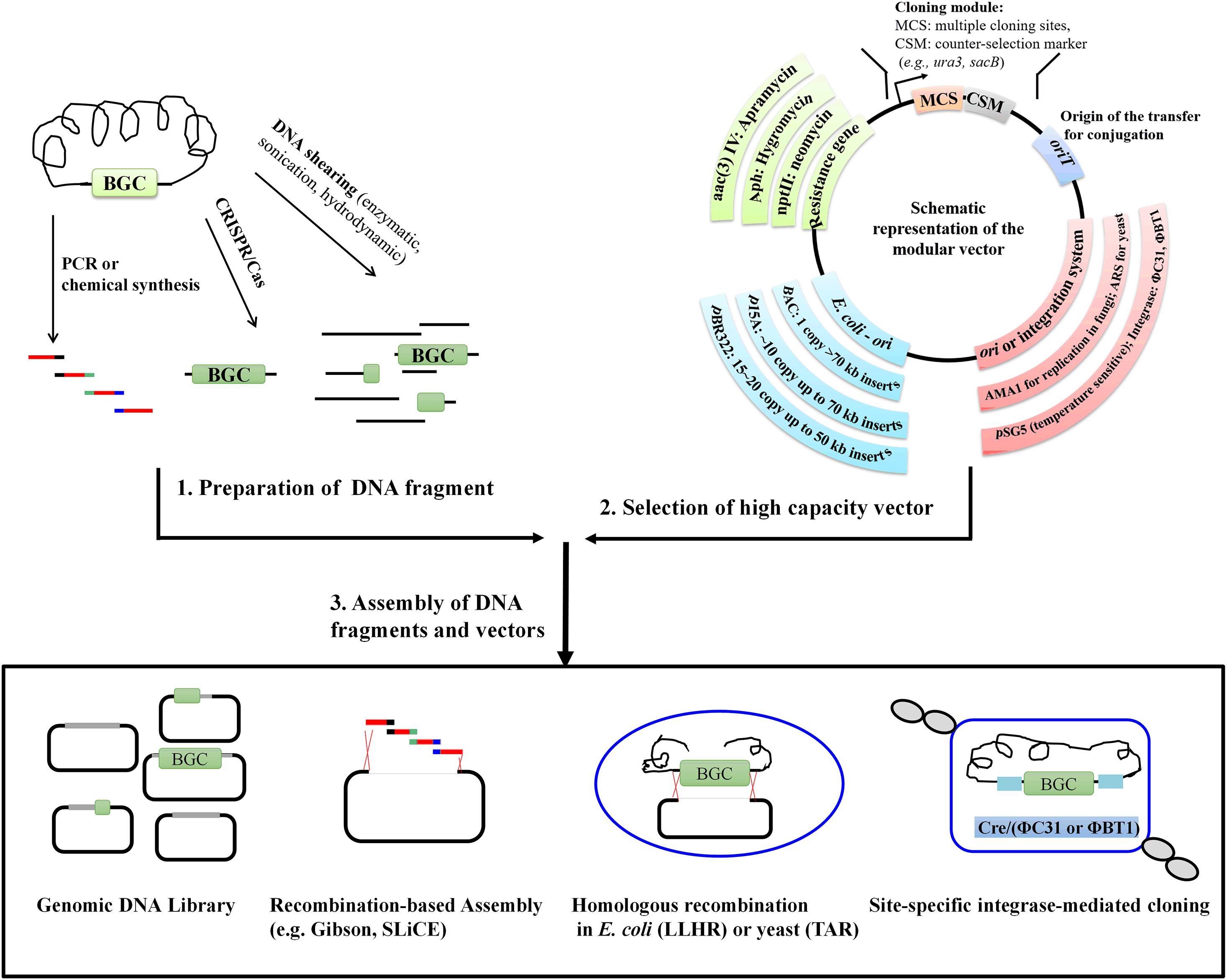
Frontiers | Recent Advances in Strategies for the Cloning of Natural Product Biosynthetic Gene Clusters
Escherichia coli β-clamp slows down DNA polymerase I dependent nick translation while accelerating ligation | PLOS ONE

A two-nuclease pathway involving RNase H1 is required for primer removal at human mitochondrial OriL. - Abstract - Europe PMC

Thermus thermophilus DNA Ligase Connects Two Fragments Having Exceptionally Short Complementary Termini at High Temperatures | Biochemistry

DNA synthesis by Klenow fragment exonuclease minus (KF exo-) on a DNA... | Download Scientific Diagram

Structure of the Herpes Simplex Virus 1 Genome: Manipulation of Nicks and Gaps Can Abrogate Infectivity and Alter the Cellular DNA Damage Response | Journal of Virology
Outline of in vivo and in vitro gap-filling assays. (A) The in vivo... | Download Scientific Diagram

The enzymatic properties of Arabidopsis thaliana DNA polymerase λ suggest a role in base excision repair | Plant Molecular Biology

An enzyme that seals gaps or breaks in double-stranded DNA is called ______. (a) DNA autosome (b) DNA polymerase (c) DNA replication (d) DNA diploid (e) DNA ligase. | Homework.Study.com

Multiplex ligase‐based genotyping methods combined with CE - Shin - 2014 - ELECTROPHORESIS - Wiley Online Library