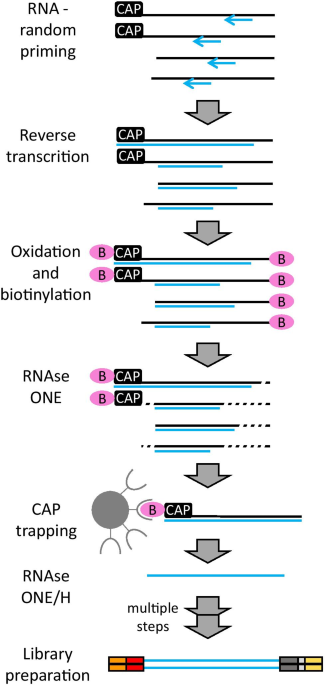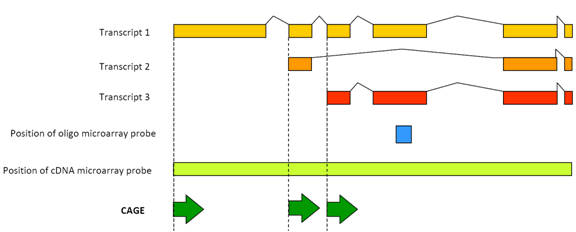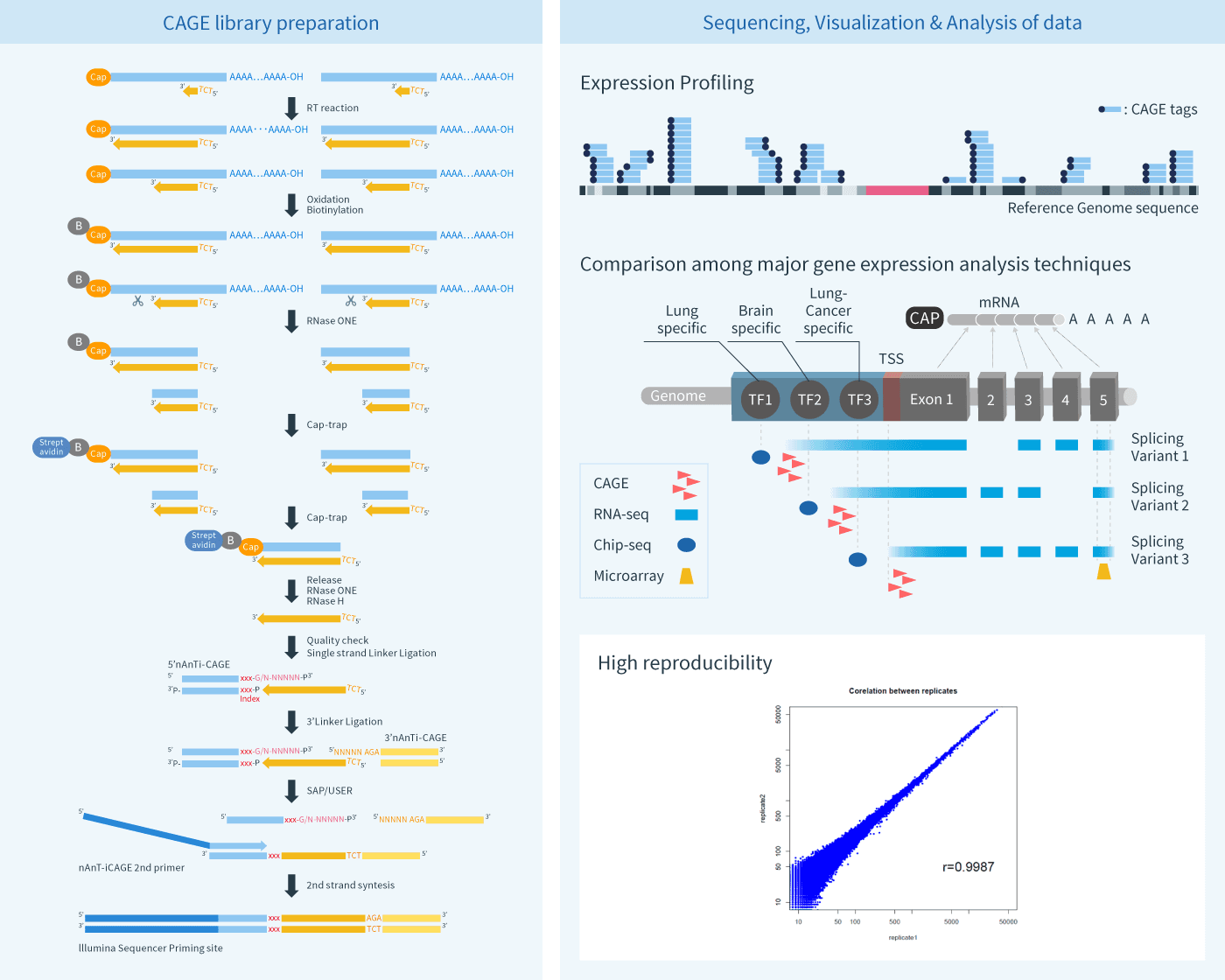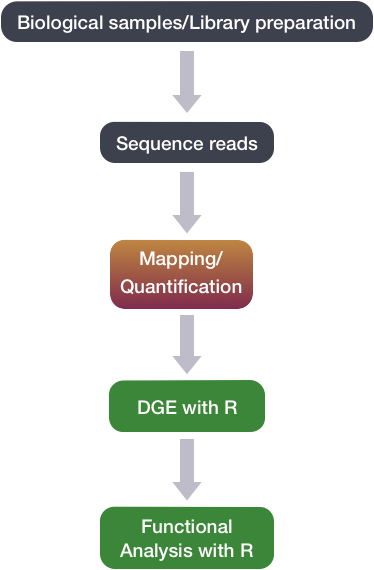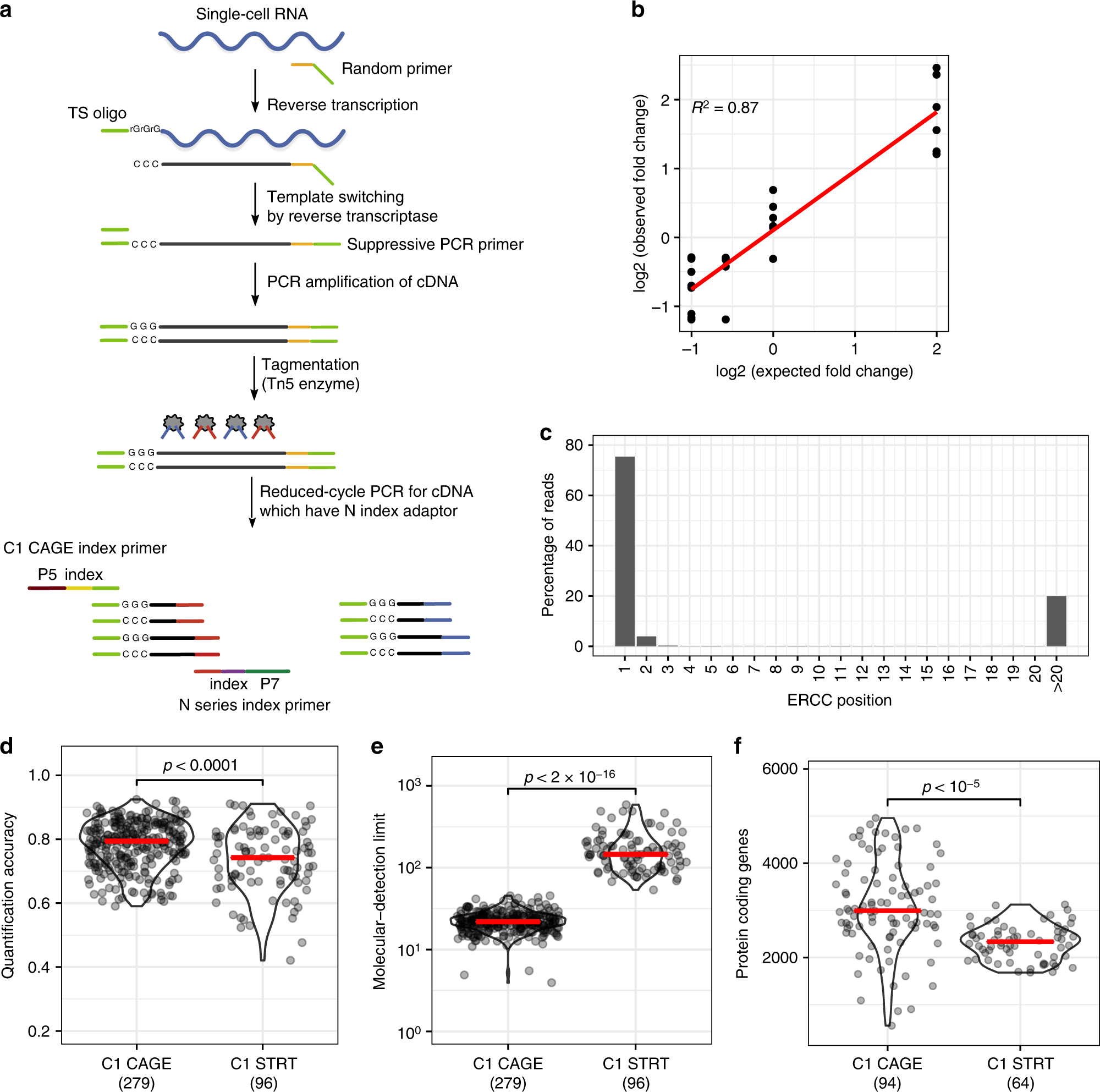
C1 CAGE detects transcription start sites and enhancer activity at single-cell resolution | Nature Communications

Simultaneous Organic and Inorganic Host‐Guest Chemistry within Pillararene‐Protein Cage Frameworks - Shaukat - 2022 - Chemistry – A European Journal - Wiley Online Library

CAGE sequencing reveals CFTR-dependent dysregulation of type I IFN signaling in activated cystic fibrosis macrophages | Science Advances
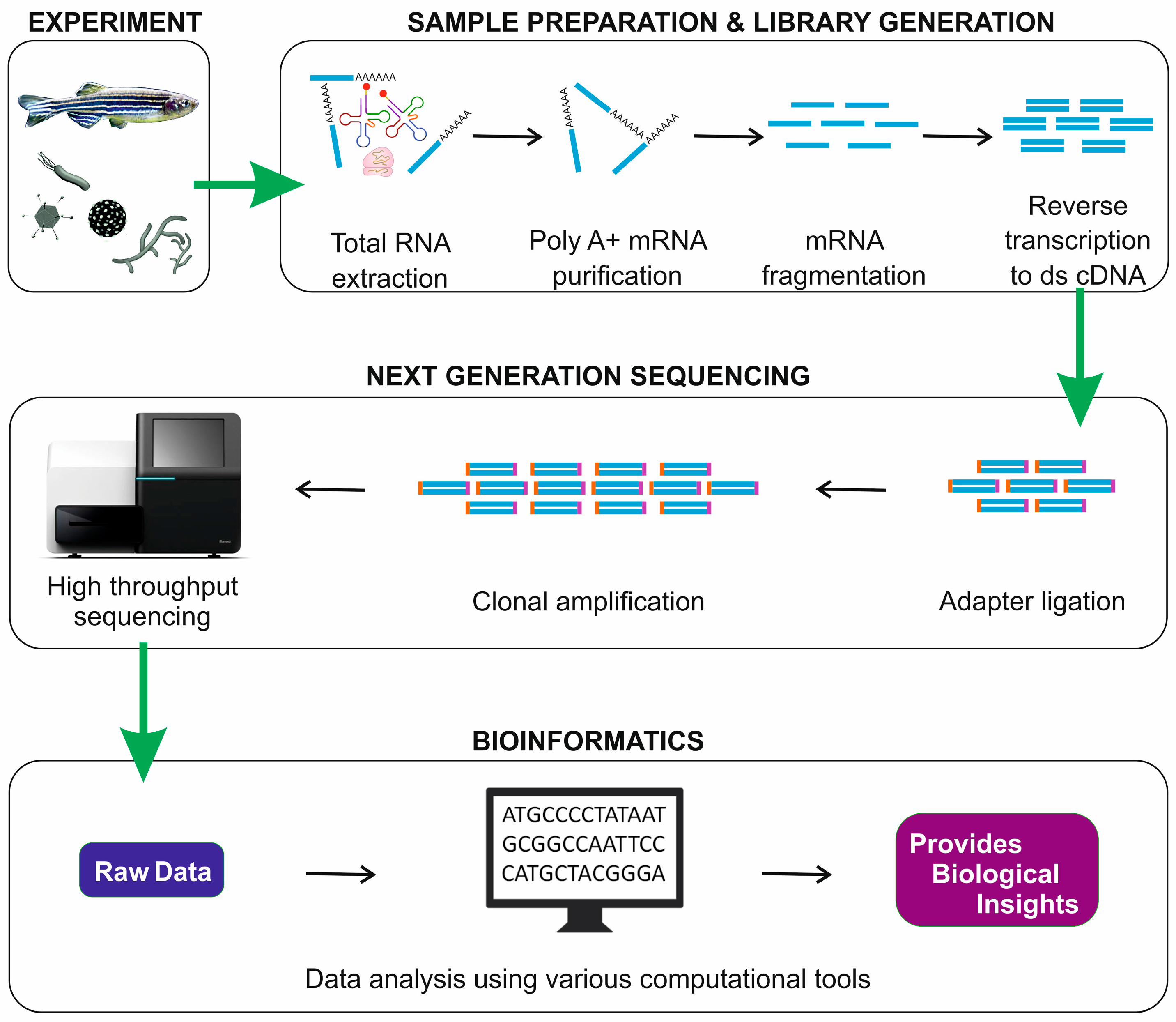
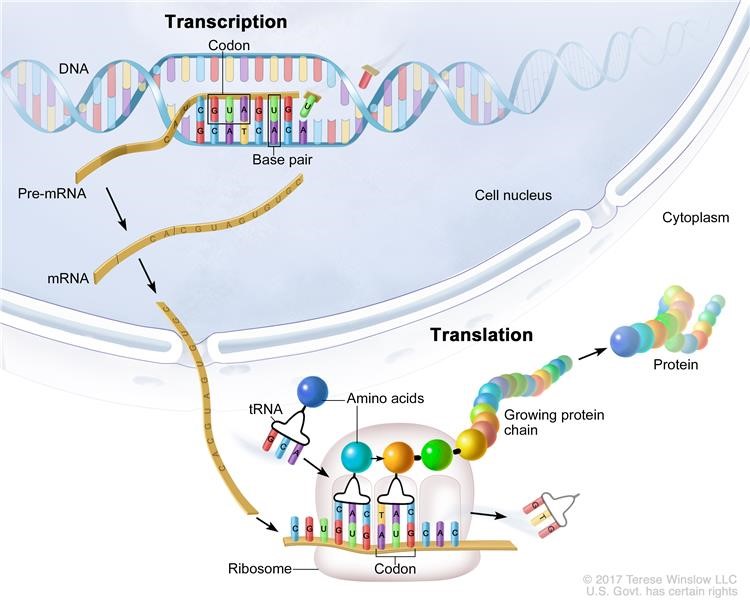
IJMS | Free Full-Text | Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review
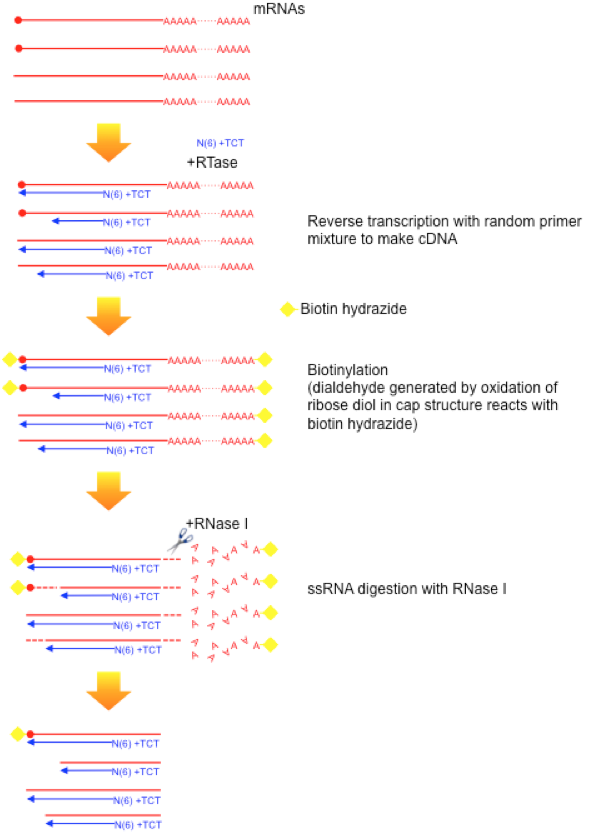
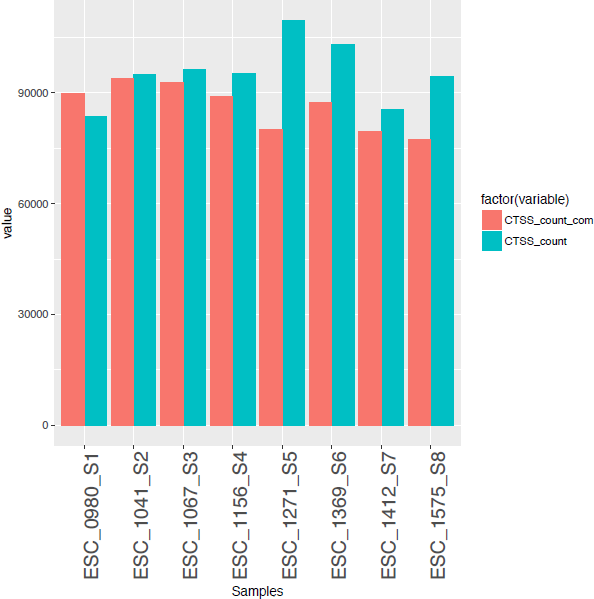
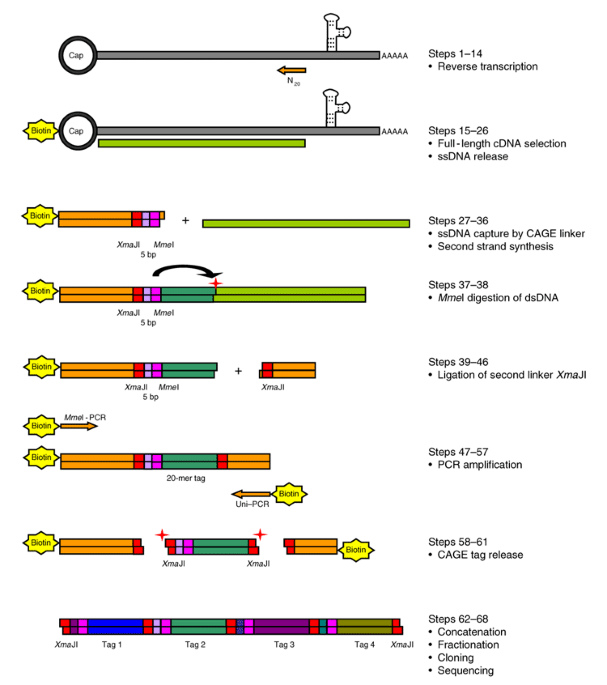
CAGE : A method for genome-wide identification of transcription start sites | 理化学研究所 ライフサイエンス技術基盤研究センター
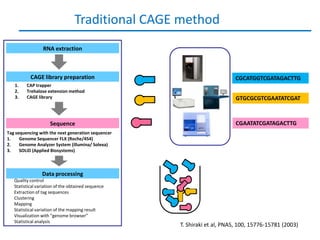
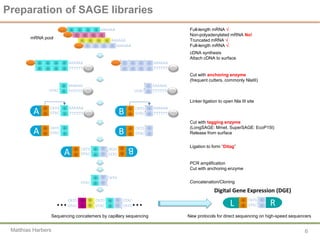
Sequencing the transcriptome reveals complex layers of regulation, Department of Biosciences and Nutrition, Karolinska Institutet, Carsten Daub Copenhagenomics 2012 | PPT

Selective translational usage of TSS and core promoters revealed by translatome sequencing | BMC Genomics | Full Text
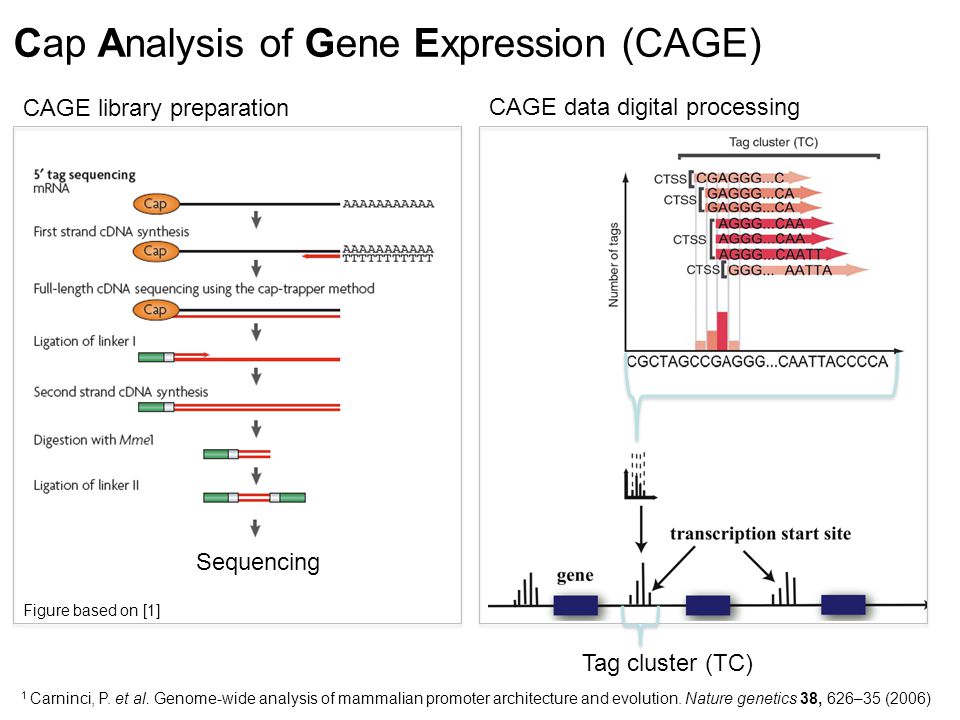
Inferring Transcriptional Regulation Using Transctiptomics Carsten O. Daub September 1 st, 2014 StratCan Summer School 2014 Vår Gård, Saltsjöbaden. - ppt download
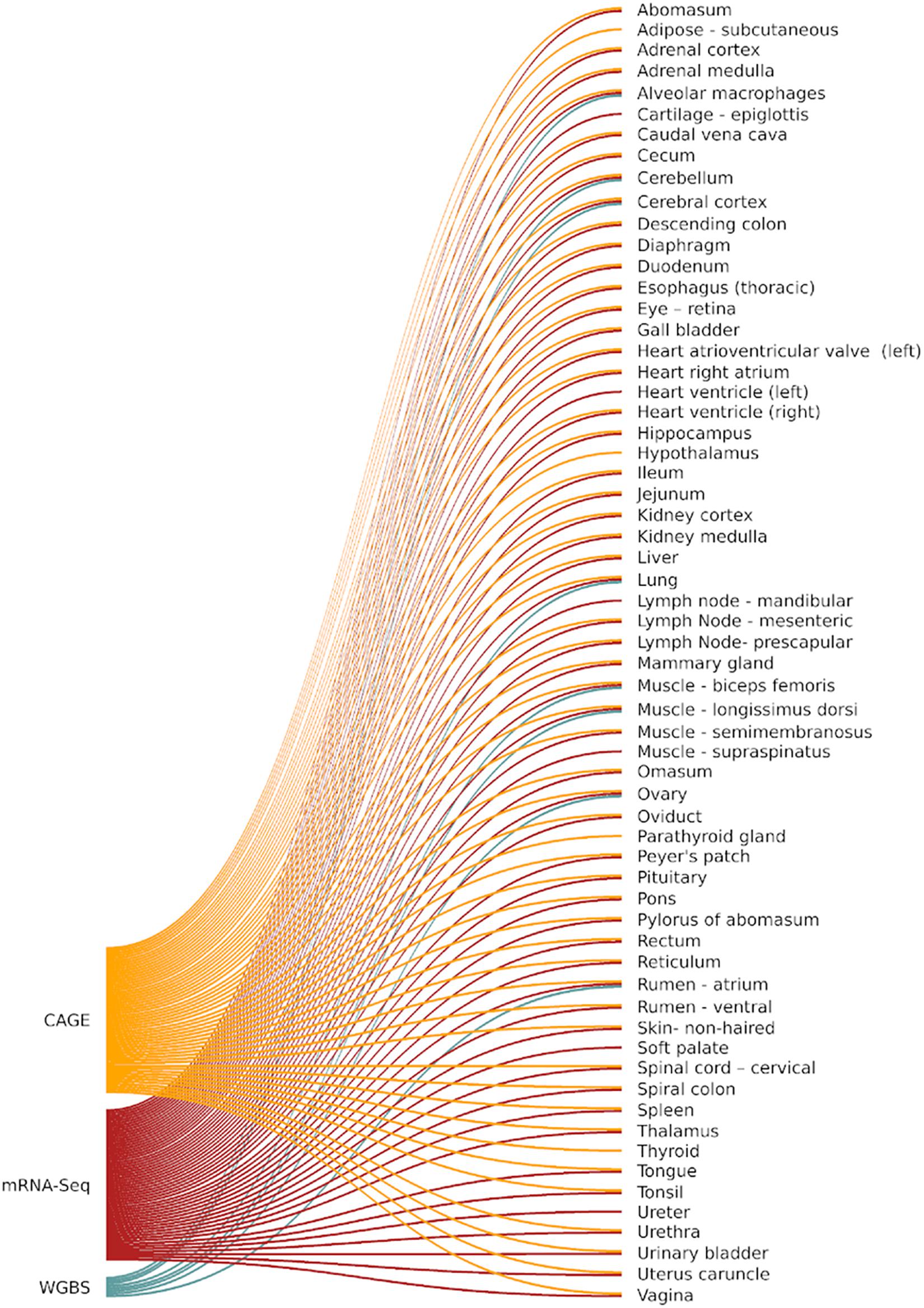
Frontiers | Global Analysis of Transcription Start Sites in the New Ovine Reference Genome (Oar rambouillet v1.0)

KSHV 3.0: a state-of-the-art annotation of the Kaposi's sarcoma-associated herpesvirus transcriptome using cross-platform sequencing | mSystems
mRNA 5′ terminal sequences drive 200-fold differences in expression through effects on synthesis, translation and decay | PLOS Genetics