A pipeline for completing bacterial genomes using in silicoand wet lab approaches | BMC Genomics | Full Text
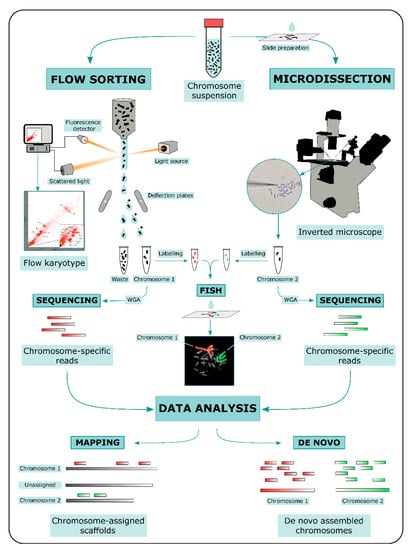
Genes | Free Full-Text | Bridging the Gap between Vertebrate Cytogenetics and Genomics with Single-Chromosome Sequencing (ChromSeq)

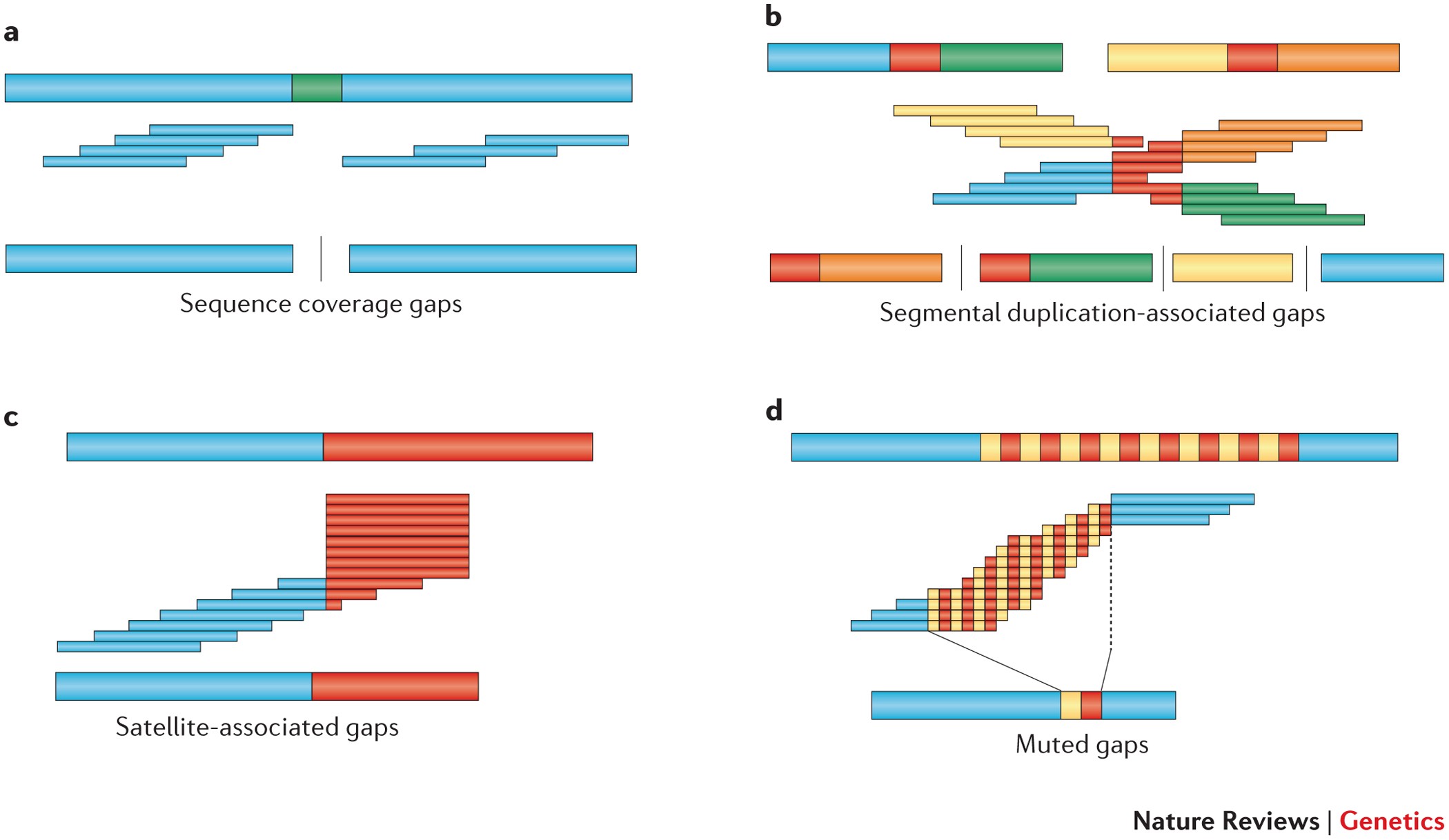
Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird‐of‐paradise - Peona - 2021 - Molecular Ecology Resources - Wiley Online Library

Whole Genome de novo Assembly |Supporting de novo assembly training |Course |WGS sequencing | data analysis| Workshop
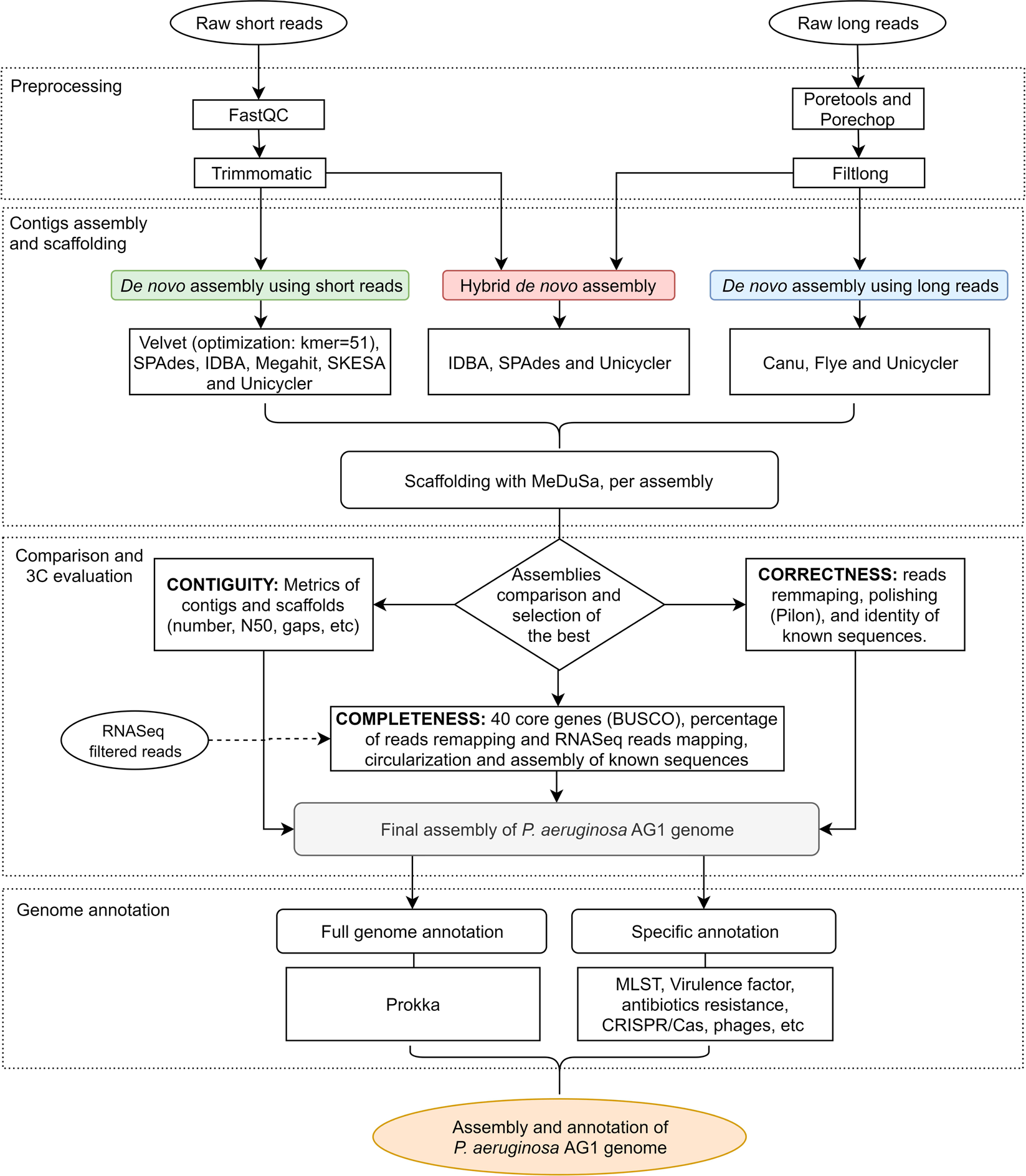
High quality 3C de novo assembly and annotation of a multidrug resistant ST-111 Pseudomonas aeruginosa genome: Benchmark of hybrid and non-hybrid assemblers | Scientific Reports

Katie Sandlin on X: "Plant genome sequence assembly in the era of long reads: Progress, challenges and future directions https://t.co/fCA7X7rpsu https://t.co/JsddDiUZBh" / X

Expanding the space of protein geometries by computational design of de novo fold families | Science
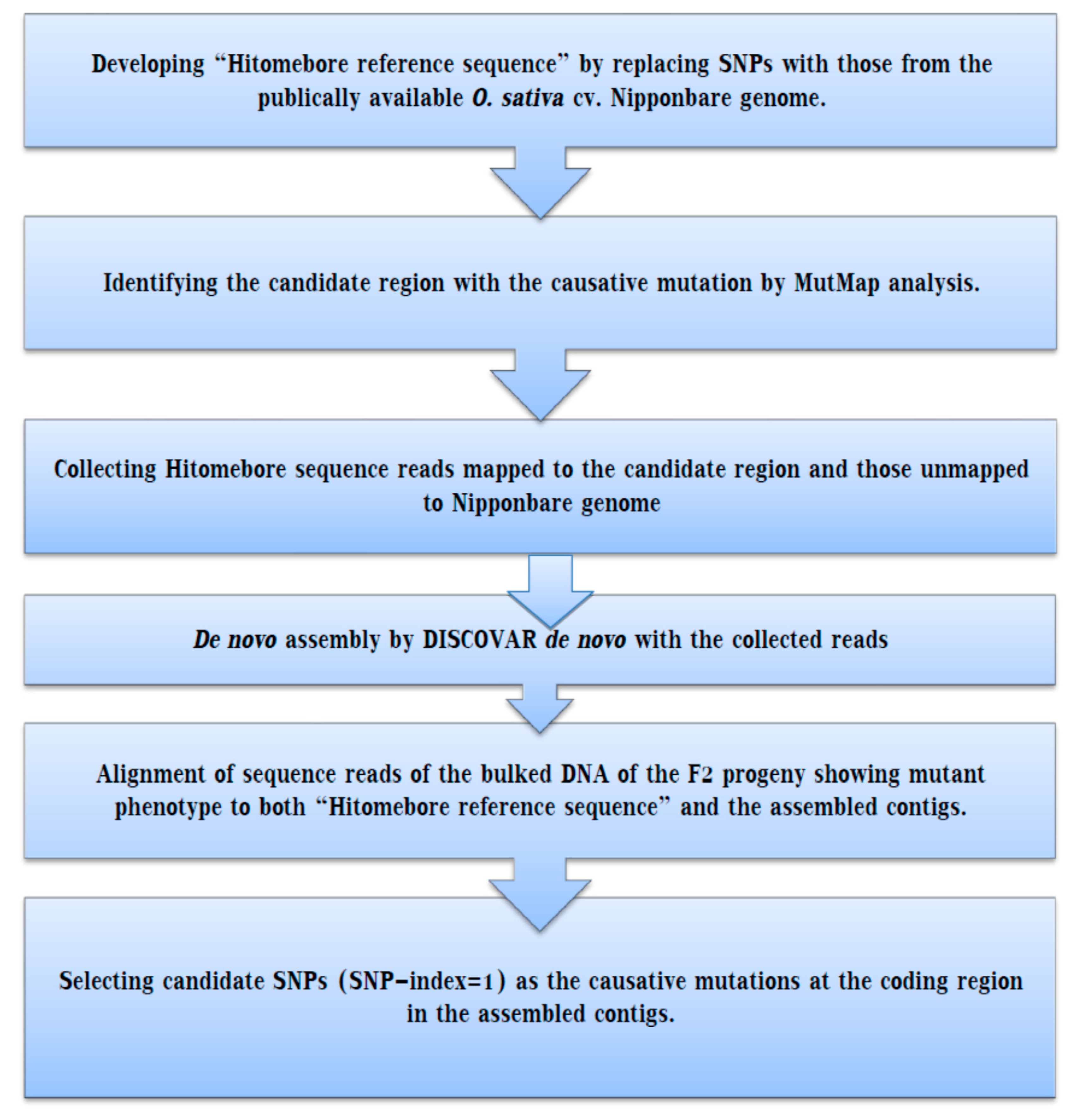
IJMS | Free Full-Text | Whole Genome Resequencing from Bulked Populations as a Rapid QTL and Gene Identification Method in Rice
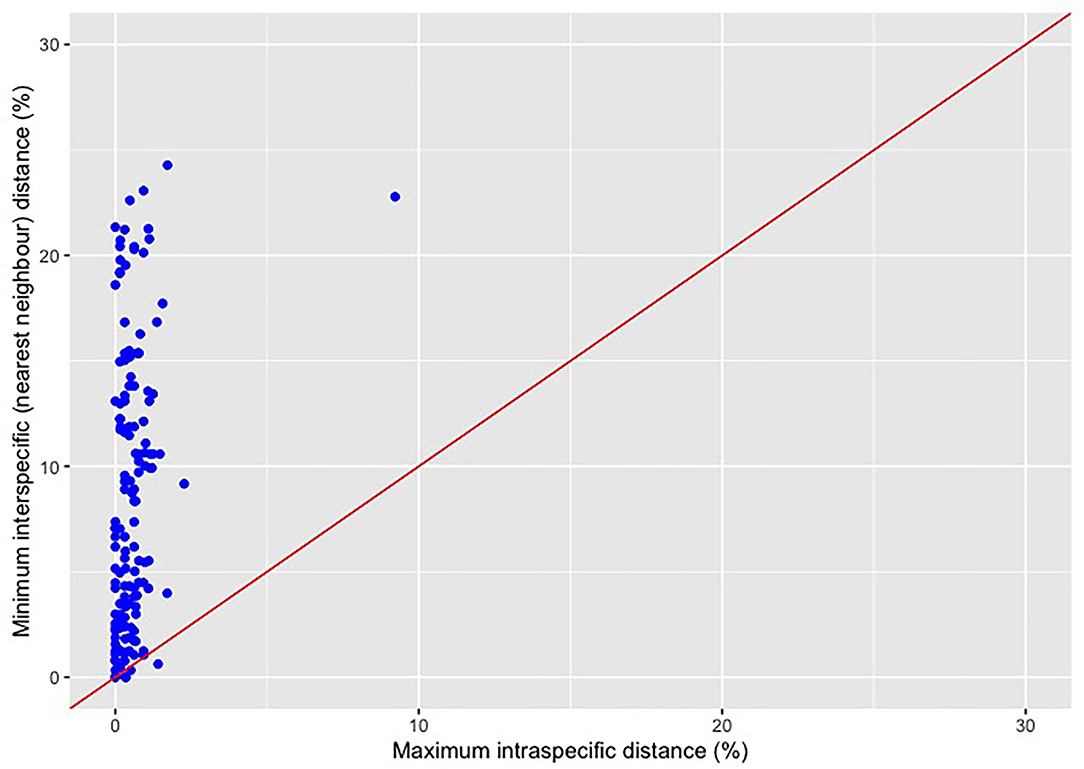
Frontiers | Lack of Statistical Rigor in DNA Barcoding Likely Invalidates the Presence of a True Species' Barcode Gap

Identifying the causes and consequences of assembly gaps using a multiplatform genome assembly of a bird‐of‐paradise - Peona - 2021 - Molecular Ecology Resources - Wiley Online Library

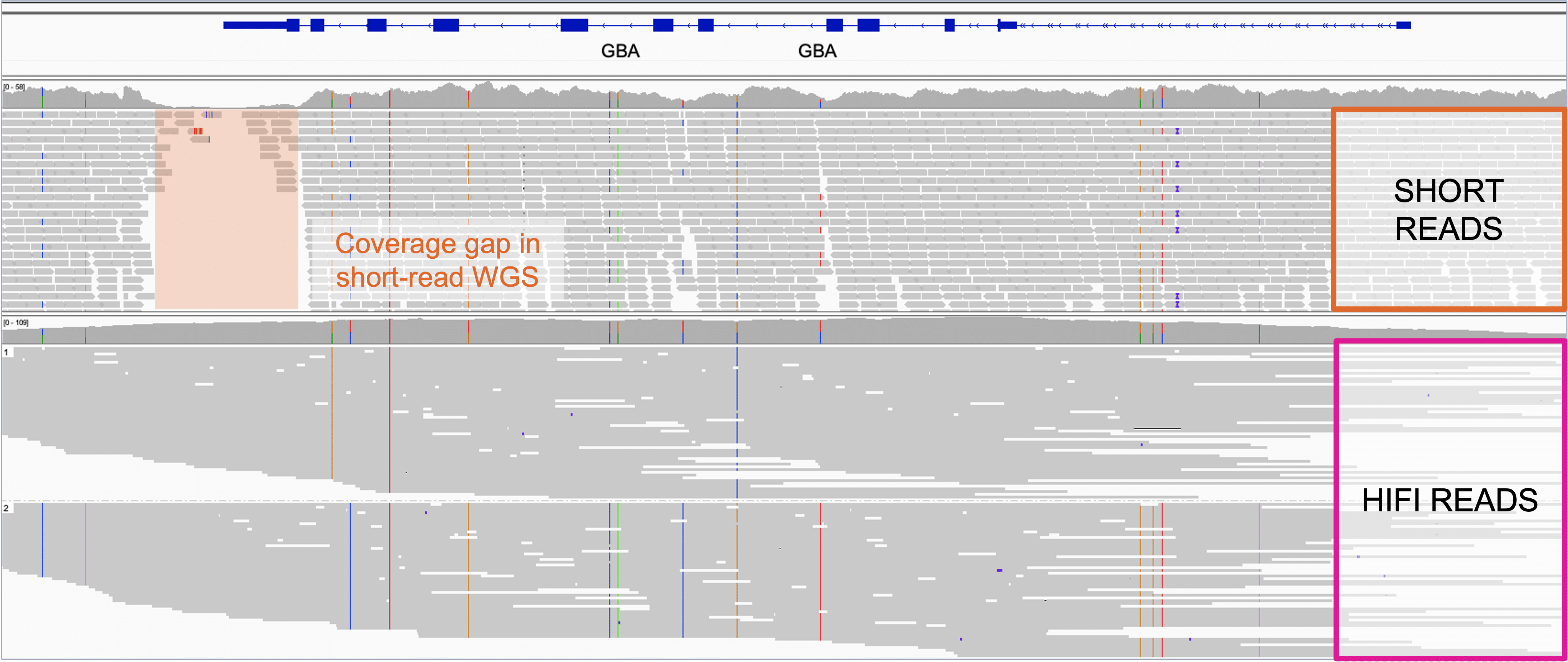
GAPPadder: a sensitive approach for closing gaps on draft genomes with short sequence reads | BMC Genomics | Full Text

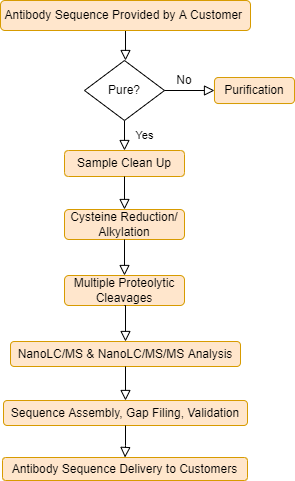
Mass Spectrometry-Based De Novo Sequencing of Monoclonal Antibodies Using Multiple Proteases and a Dual Fragmentation Scheme | Journal of Proteome Research

Integrated gene analyses of de novo variants from 46,612 trios with autism and developmental disorders | PNAS

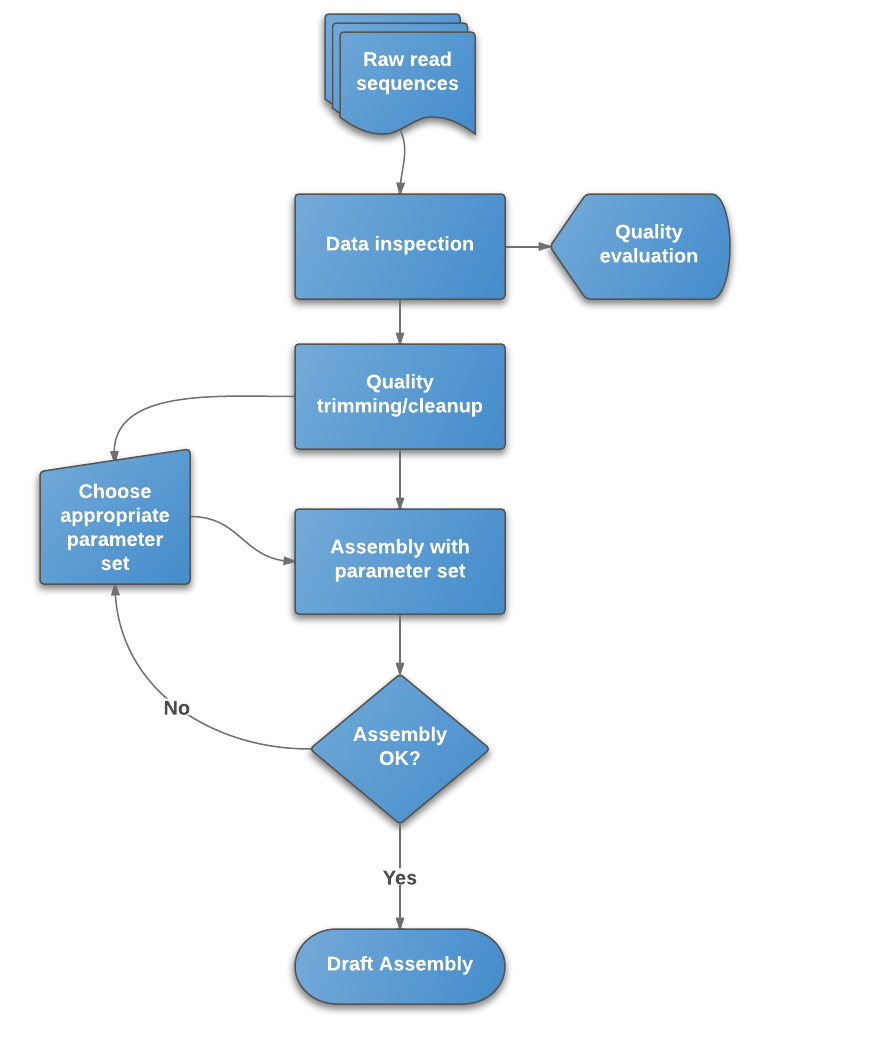
De novo assembly. High-level diagram of long-read assembly pipeline.... | Download Scientific Diagram
Selecting Superior De Novo Transcriptome Assemblies: Lessons Learned by Leveraging the Best Plant Genome | PLOS ONE